Authors
Feng Zhang
Summary
Below is a concise protocol for cloning CRISPR-Cas9 genome editing constructs.
Introduction
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is a microbial nuclease system involved in defense against invading phages and plasmids. CRISPR loci in microbial hosts contain a combination of CRISPR-associated (Cas) genes as well as non-coding RNA elements capable of programming the specificity of the CRISPR-mediated nucleic acid cleavage. We have recently harnessed the type II CRISPR nuclease system to facilitate genome editing in mammalian cells (Cong et al., Science 2013).
The CRISPR/Cas system can be implemented in mammalian cells by co-expressing the bacterial Cas9 nuclease along with the guide RNA. Two forms of guide RNAs can be used to facilitate Cas9-mediated genome cleavage, using a CRISPR RNA array and tracrRNA or a synthetic guide RNA fusing the CRISPR RNA with the tracrRNA. These two systems are described below.
Materials
1. S. pyogenes Cas9 (or Cas9 D10A nickase) + CRISPR RNA array + tracrRNA: This plasmid contains three expression cassettes. In order to target a given site, the plasmid can be digested using BbsI, and a pair of annealed oligos (design is indicated below) can be cloned into the CRISPR array. The oligos is designed based on the target site sequence (30bp) and needs to be flanked on the 3′ end by a 3bp NGG PAM sequence.
Genbank Map of Backbone Plasmid PX260 (rev. 20130212)
Genbank Map of Backbone Plasmid PX334 (rev. 20130212)
2. S. pyogenes Cas9 (or Cas9 D10A nickase) + chimeric guide RNA containing +85nt of tracrRNA: This plasmid contains two expression cassettes, hSpCas9 and the chimeric guide RNA. The vector can be digested using BbsI, and a pair of annealed oligos (design is indicated below) can be cloned into the guide RNA. The oligos is designed based on the target site sequence (20bp) and needs to be flanked on the 3′ end by a 3bp NGG PAM sequence. We have found that increasing the length of the chimeric guide RNA can increase targeting efficiency; therefore this version of the backbone contains a longer fragment of the tracrRNA (+85nt).
Design note for chimeric backbones: Please note that for PX330/335 cloning backbone, within the guide sequence insert, the genome target was illustrated above as ‘GNNNNNNNNNNNNNNNNNNN’, i.e., one base ‘G’ followed by 19 Ns, instead of the 20 Ns shown for the PX260/334 cloning backbones. This difference in oligo design was due to the requirement of human U6 promoter to have a ‘G’ base at the transcription start site. Hence, we recommend finding a 20bp genome target starting with the base ‘G’. If you have to use other bases at the starting position of your genome target, you could add an additional ‘G’ to the front of your target, as described in our FAQs and Trouble Shooting Tips page.
Genbank Map of Backbone Plasmid PX330 (rev. 20130212)
Genbank Map for Backbone Plasmid PX335 (rev. 20130212)
References
Multiplex Genome Engineering using CRISPR/Cas Systems
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F.
Science. 2013 Jan 3. DOI: 10.1126/science.1231143
Documents for download
Stats
- Recommendations +1 100% positive of 1 vote(s)
- Views 2539
- Comments 0
Recommended
-

Julian Ji
Main position not set
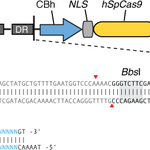 Image 1
Image 1 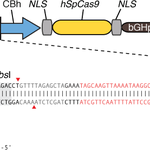 Image 2
Image 2